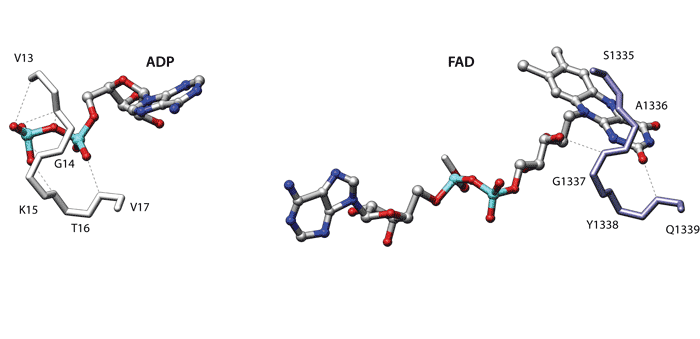
The Figure displays the similarity between dethiobiotin synthetase from Escherichia coli (1dak, left) and D-amino acid oxidase from the yeast Rhodotorula gracilis (1c0i, right). When comparing the binding sites of these two proteins superpose3D finds a structural match involving the C-alphas of eight different residues which can be superimposed with an RMSD of 0.66 Å.
Dethibiotin synthetase belongs to the extensive group of nucleoside triphosphate hydrolases containing the characteristic phosphate-binding P-loop. Conversely the D-amino acid oxidase belongs to the “Nucleotide-binding domain” fold, which is a member of the large group of “Rossmann-like” folds. This protein binds the O2 and O3’ of the riboflavine moiety using a loop which is very similar to the P-loop of Dethibiotin synthetase.
These compact loops often bind anions with hydrogen bonds to main chain atoms and have been termed “nests” [Watson JD, Milner-White EJ, J Mol Biol. (2002) 315(2):171-82]. Given this mode of binding the identity of the residues and the position of the side chain is not important in this case. Accordingly four of the five residues that comprise the loops have negative substitution scores in a BLOSUM62 matrix.

