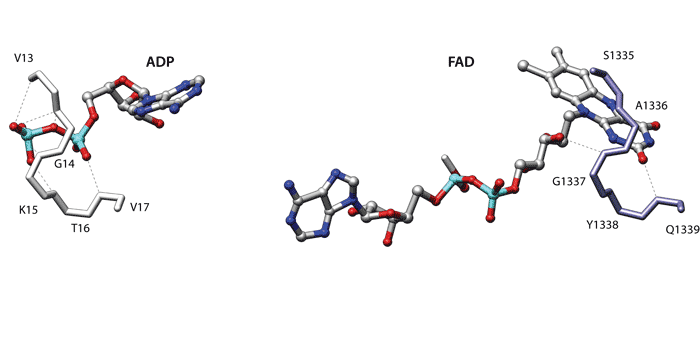
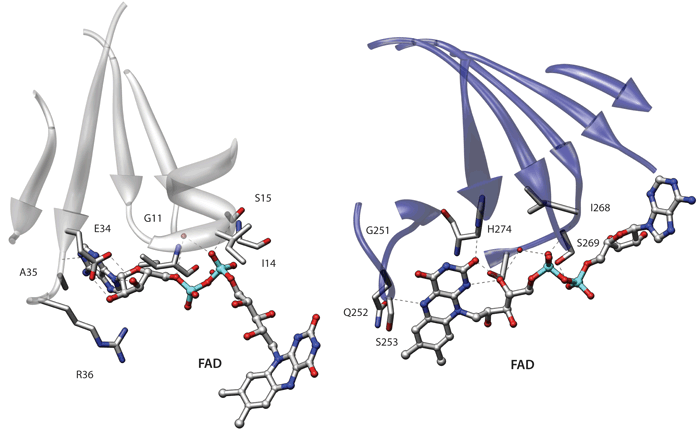
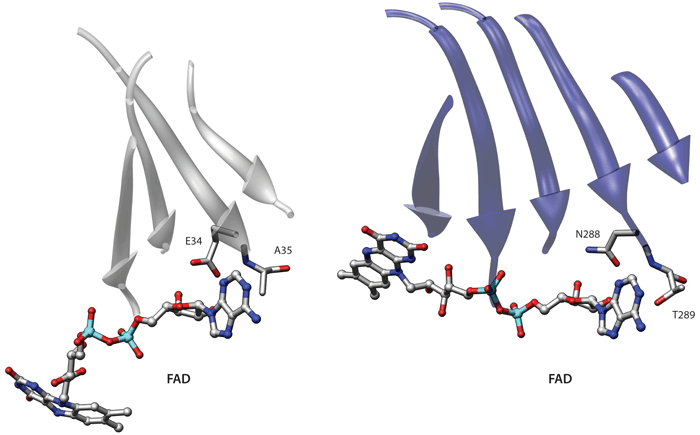
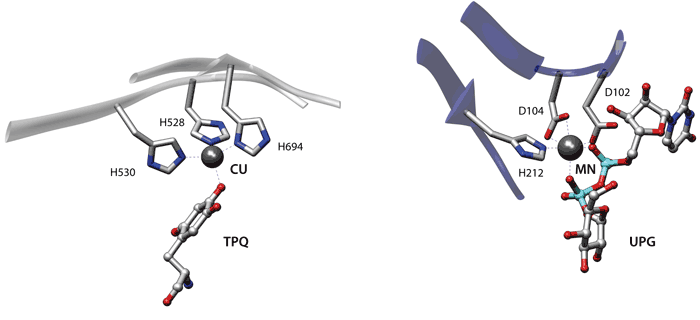
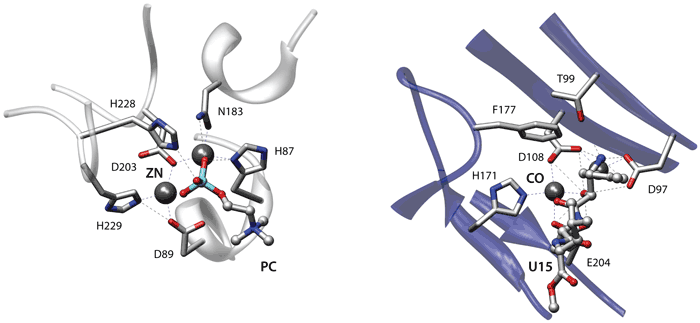
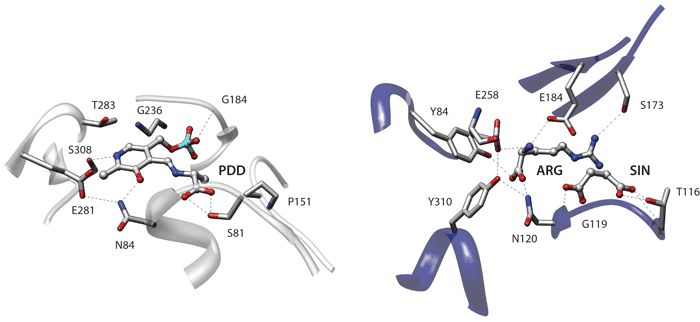
For instance if you are only interested in the overall shape of a functional patch you may choose to only consider the position of the C-alpha. Alternatively, if you want to compare hydrogen bonding pattern in a binding site or enzyme active site, you need a more detailed representation that includes information about the side chain groups.
This five examples demonstrate the results that can be obtained with different parameter settings. Remember that these are just examples, any kind of representation is possible! We choose ligand binding sites as a test case because the position of the ligands immediately tells whether the similarity has a functional significance. However any kind of functional patch (e.g. enzyme active sites, protein/protein interaction surfaces etc.) can be analyzed with superpose3D. These examples are discussed in detail in the superpose3D paper (see References)
The first two examples use a coarse description that only considers the position of the C-alpha residues. In the third example each residue is represented with the C-alpha and the geometric centroid of the side chain residues. Finally the last two examples use a more elaborate description focused on the specific side-chain chemical groups of each residue. Click on each image to go to a detailed description of the structures or on the titles to view an explanation of the parameters.
An archive with all the necessary files to reproduce these examples is available at the Downloads page.
Coarse description (C-alpha only)
Including side chain information (C-alpha + geometric centroid of side chain)
Detailed representation (specific chemical groups)