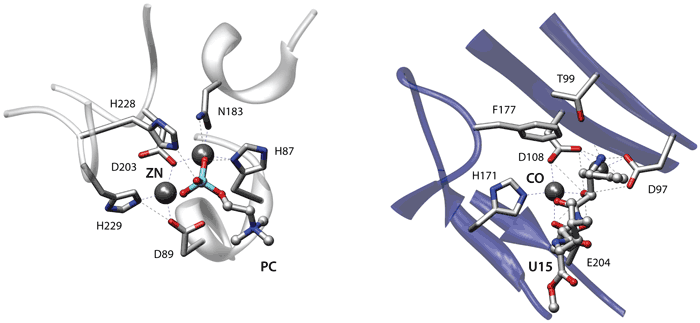
The figure shows an alignment of the active sites of the teichoic acid phosphorylcholine esterase Pce from Streptococcus pneumoniae (2bib, left) and of a methionine aminopeptidase from Escherichia coli (2gg8, right). For both enzymes a mechanism has been proposed whereby the two metal ions in the active site activate a water molecule for nucleophilic attack and participate in the stabilization of the resulting tetrahedral intermediate [Hermoso JA et al., Nat Struct Mol Biol. (2005) 12:533-8; Lowther WT et al., Biochemistry (1999) 38:14810-9].
The two proteins are unrelated and belong to different SCOP folds. When comparing the binding sites of these proteins superpose3D identified a match comprising eight pseudoatoms belonging to six residues with an RMSD of 0.68 Å. Interestingly the phosphorylcholine esterase is complexed with its phosphocholine substrate while the methionine aminopeptidase is bound to an aminoacidic inhibitor.
The algorithm therefore succeeded both in identifying a similarity between two unrelated enzymes that share similar mechanisms and also in highlighting the similar binding modes of the substrate and an inhibitor. The representation focused on chemical groups was absolutely necessary to identify the structural similarity. Indeed the metal binding residues Asp97, Asp108, His171 of the aminopeptidase (2gg8) and His87, Asp203, His229 of the phosphorylcholine esterase (2bib) only have the chemical groups that are involved in the interaction superimposed while the remaining atoms occupy different spatial positions.

