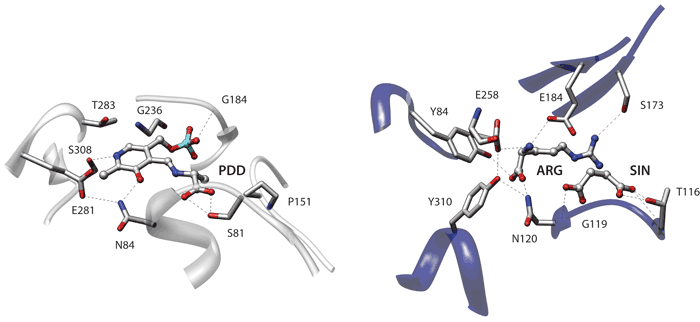
The figure depicts the similarity between the binding sites of Serine racemase from Schizosaccharomyces pombe (2zpu, left) and Argininosuccinate synthetase from Thermus thermophilus (1kor, right). These proteins belong to different PFAM families and have very low sequence identity (20%). They also bind completely different ligands. Argininosuccinate synthetase is complexed with arginine and succinate while serine racemase is covalently bound with a modified Pyridoxal phosphate moiety (PLP-D-Ala). Superpose3D identifies a similarity comprising nine pseudoatoms belonging to eight residues with an RMSD of 0.64 Å. Interestingly the structural match overlays the ligands so that arginine and succinate are superimposed with different parts of the PLP-D-Ala molecule (see the figure below where the matching pseudoatoms are represented as spheres).
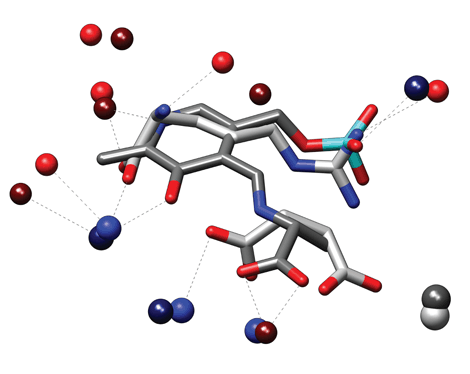
Arginine is superimposed to the pyridoxal phosphate moiety, with the guanidinium group in the same position as the phosphate of PLP. Succinate occupies a position corresponding to the alanine moiety of PLP-D-Ala. Again the identification of this similarity was possible because a residue description focused on chemical groups was used. For instance Tyr84 of the Argininosuccinate synthetase and Ser308 of the Serine racemase, which are both hydrogen bonded to their respective ligands, only have their terminal oxydrile superimposed while the remaining atoms occupy completely different positions.

